Newer
Older
<img align="right" src="https://raw.githubusercontent.com/FZJ-INM1-BDA/siibra-explorer/master/docs/images/siibra-explorer-square.jpeg" width="200">
[](https://zenodo.org/badge/latestdoi/109723444)
[](https://siibra-explorer.readthedocs.io/)
[](https://opensource.org/licenses/Apache-2.0)
# siibra-explorer - Interactive atlas viewer for multilevel brain atlases
*Authors: Big Data Analytics Group, Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich GmbH*
Copyright 2020-2021, Forschungszentrum Jülich GmbH
`siibra-explorer` is a browser based 3D viewer for exploring brain atlases that cover different spatial resolutions and modalities. It is built around an interactive 3D view of the brain displaying a unique selection of detailed templates and parcellation maps for the human, macaque, rat or mouse brain, including BigBrain as a microscopic resolution human brain model at its full resolution of 20 micrometers.
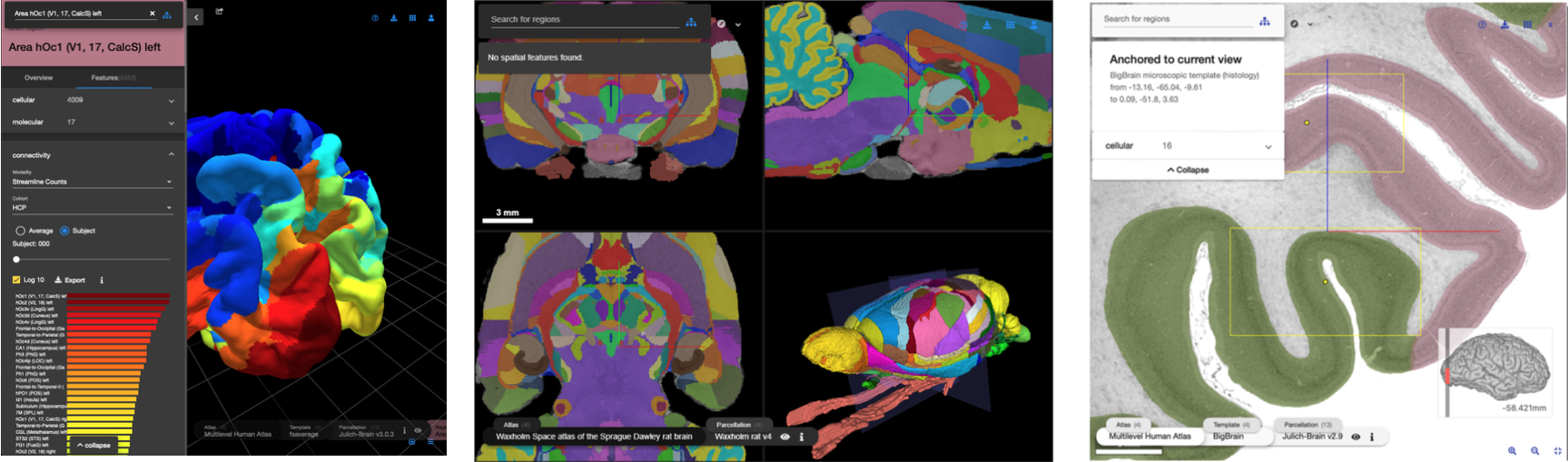
`siibra-explorer` builds on top [nehuba](https://github.com/HumanBrainProject/nehuba) for the visualization volumetric brain volumes at possible high resolutions, and [three-surfer](https://github.com/xgui3783/three-surfer) for the visualization of surface based atlases. By connecting to [siibra-api](https://github.com/fzj-inm1-bda/siibra-api), `siibra-explorer` gains access to brain atlases of different species, including to navigate their brain region hierarchies, maps in different coordinate spaces, and linked regional data features. It provides metadata integration with the [EBRAINS knowledge graph](https://kg.ebrains.eu), different forms of data visualisation, and a structured plugin system for implementing custom extensions.
A live version of the siibra explorer is available at [https://atlases.ebrains.eu/viewer/](https://atlases.ebrains.eu/viewer/). User documentation can be found at <https://siibra-explorer.readthedocs.io/>. This README.md is aimed at developers who would like to develop and run `siibra-explorer` locally.
Siibra explorer is built with [Angular (v14.0)](https://angular.io/), [Bootstrap (v4)](http://getbootstrap.com/), and [fontawesome icons](https://fontawesome.com/). Some other notable packages used are [ngrx/store](https://github.com/ngrx/platform) for state management.
Releases newer than [v0.2.9](https://github.com/fzj-inm1-bda/siibra-explorer/releases/tag/v0.2.9) also uses a nodejs backend, which uses [passportjs](http://www.passportjs.org/) for user authentication, [express](https://expressjs.com/) as the http framework.
Releases newer than [v2.13.0](https://github.com/fzj-inm1-bda/siibra-explorer/releases/tagv2.13.0) uses a python backend, which uses [authlib](https://pypi.org/project/Authlib/) for user authentication, [fastapi](https://pypi.org/project/fastapi/) as the http framework.
Development environments are stored under `src/environments/environment.common.ts`. At build time, `src/environments/environment.prod.ts` will be used to overwrite the environment.
Whilst this approach adds some complexity to the development/build process, it enhances developer experience by allowing the static typing of environment variables.
##### Buildtime environments
Please see [build_env.md](build_env.md)
##### Deploy environments
Please see [deploy_env.md](deploy_env.md)
Please see [e2e_env.md](e2e_env.md)
$ git clone https://github.com/FZJ-INM1-BDA/siibra-explorer
$ cd siibra-explorer
$ npm start
```
To run backend dev server:
```bash
$ cd backend
$ pip install -r requirements.txt
$ uvicorn app.app:app --host 0.0.0.0 --port 8080
Please see [src/plugin/README.md](src/plugin/README.md)
## Contributing
Feel free to raise an issue in this repo and/or file a PR.
## Versioning
Commit history prior to v0.2.0 is available in the [legacy-v0.2.0](https://github.com/FZJ-INM1-BDA/siibra-explorer/tree/legacy-v0.2.0) branch. The repo was rewritten to remove its dependency on neuroglancer and nehuba. This allowed for simpler webpack config, faster build time and AOT compilation.